I learned the simple concept of animation back in school, when some of my classmates would draw stick figures on the edge of large textbooks. At first I was wondering why one would defile a textbook in such a way, but then as they flipped through the pages and brought the stick figures to life, I was in awe. Despite this, at that stage of my life, a textbook was sacred to me (they were expensive and scarce), so I would use large Post-it notes to doodle instead. I wasn't very good at drawing (even when it comes to stick figures), so I made a few animations and that was it.
This post is on creating animated plots using R. I wrote it not because I wanted to rekindle my youthful interest in stick figure animation but because I wanted to create an animated plot for an upcoming talk. I found a short post on creating animated plots using R and I follow the same idea of making multiple plots and then combining them into a GIF using ImageMagick.
#number of frames or plots
frames <- 50
# function for creating file name with leading zeros
# makes it easier to process them sequentially
rename <- function(x){
if (x < 10) {
return(name <- paste('000',i,'plot.png',sep=''))
}
if (x < 100 && i >= 10) {
return(name <- paste('00',i,'plot.png', sep=''))
}
if (x >= 100) {
return(name <- paste('0', i,'plot.png', sep=''))
}
}
#loop through plots
for(i in 1:frames){
name <- rename(i)
#saves the plot as a .png file in the working directory
png(name)
sd <- 10
n <- 10000
factor <- i * 2
m <- 50 + factor
x <- rnorm(n, m, sd)
hist(x,
xlim=c(0,200),
ylim=c(0,2000),
main = paste('Histogram of rnorm() n = ', n, ' mean = ', m, ' sd = ', sd),
)
dev.off()
}
#run ImageMagick
my_command <- 'convert *.png -delay 3 -loop 0 animation.gif'
system(my_command)
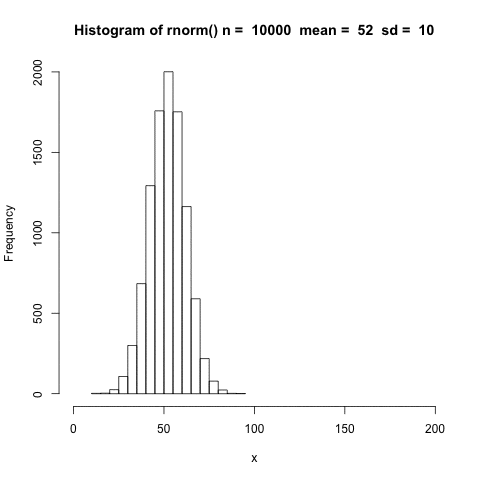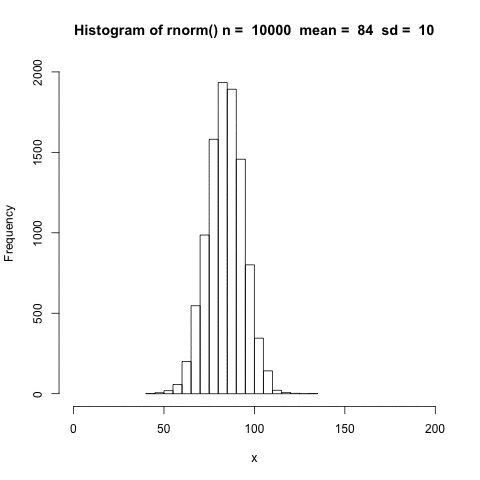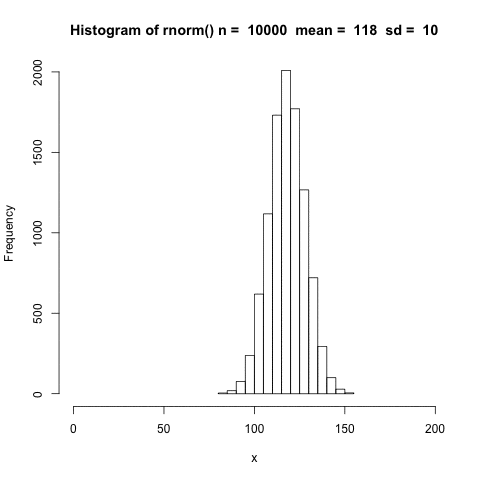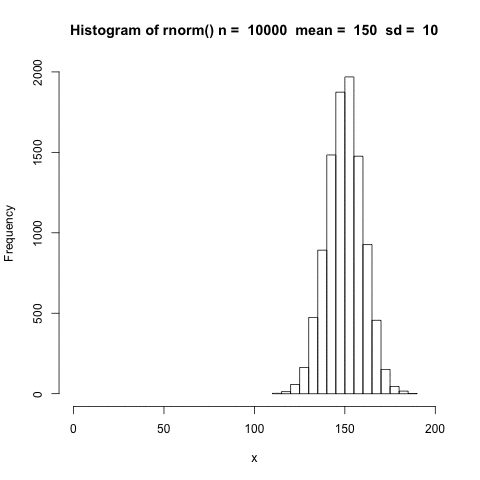 The distribution shifts according to the mean.
The distribution shifts according to the mean.
Visualise filtering threshold
A practical use of animated plots could be to visualise the effects of independent filtering on the number of genes detected as differentially expressed.
frames <- 50
rename <- function(x){
if (x < 10) {
return(name <- paste('000',i,'plot.png',sep=''))
}
if (x < 100 && i >= 10) {
return(name <- paste('00',i,'plot.png', sep=''))
}
if (x >= 100) {
return(name <- paste('0', i,'plot.png', sep=''))
}
}
#I host this file on my server for convenience
file_url <- 'http://davetang.org/eg/pnas_expression.txt'
data <- read.table(file_url,
header=T,
sep="\t",
stringsAsFactors=F,
row.names=1
)
#remove length column
data <- data[,-8]
library(edgeR)
#loop through plots
for(i in 1:frames){
name <- rename(i)
#saves the plot as a .png file in the working directory
png(name)
data_subset <- subset(data, rowSums(data)>i)
group <- c(rep("Control",4), rep("Test",3))
d <- DGEList(counts = data_subset, group=group)
#data normalisation
d <- calcNormFactors(d, method="TMM")
d <- estimateDisp(d)
et <- exactTest(d)
de <- table(p.adjust(et$table$PValue, method = "BH")<0.05)[2]
plot(et$table$logCPM,
et$table$logFC,
ylim=c(-10,10),
xlim=c(0,14),
pch=19,
cex=0.1,
col=as.numeric(p.adjust(et$table$PValue, method = "BH")<0.05)+1,
main=paste("Independent filtering at", i, ';', de, 'differentially expressed')
)
dev.off()
}
#run ImageMagick
#I slowed it down a bit more
my_command <- 'convert *.png -delay 5 -loop 0 filtering.gif'
system(my_command)
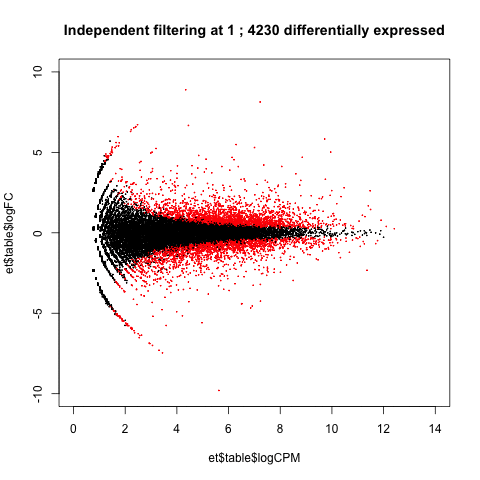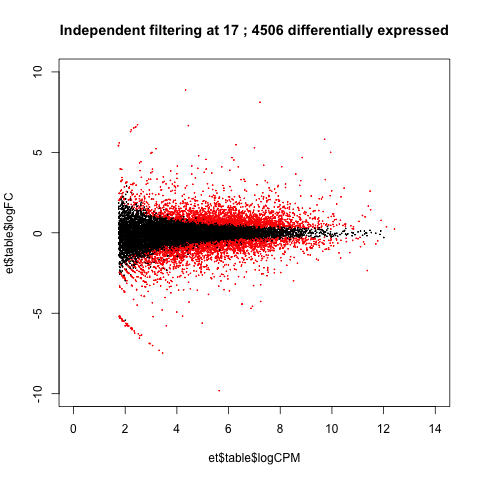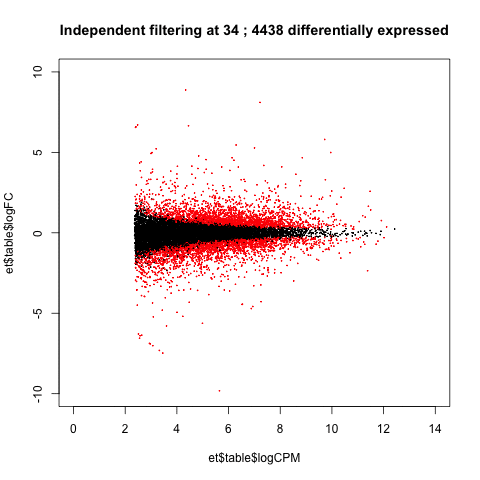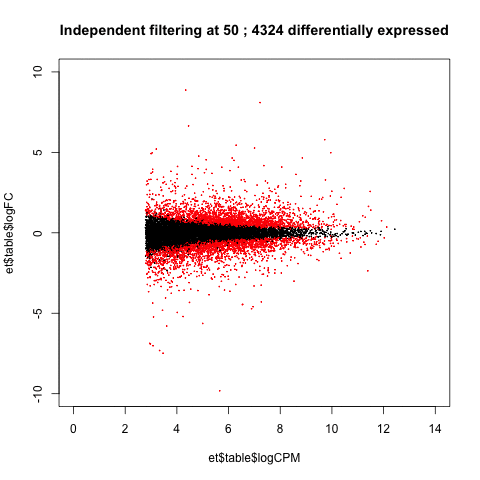 Adjusting the filtering threshold did not have a large effect on the number of differentially expressed genes. (This isn't always the case.)
Adjusting the filtering threshold did not have a large effect on the number of differentially expressed genes. (This isn't always the case.)
Rotating 3D scatter plot
#install.packages("scatterplot3d")
library(scatterplot3d)
data(iris)
frames <- 360
rename <- function(x){
if (x < 10) {
return(name <- paste('000',i,'plot.png',sep=''))
}
if (x < 100 && i >= 10) {
return(name <- paste('00',i,'plot.png', sep=''))
}
if (x >= 100) {
return(name <- paste('0', i,'plot.png', sep=''))
}
}
p <- prcomp(iris[,1:4])
my_col <- as.numeric(iris$Species)
#loop through plots
for(i in 1:frames){
name <- rename(i)
#saves the plot as a .png file in the working directory
png(name)
scatterplot3d(p$x[,1:3],
main=paste("Angle", i),
angle=i,
pch=19,
cex.symbols=0.5,
color=my_col)
dev.off()
}
my_command <- 'convert *.png -delay 1 -loop 0 3d.gif'
system(my_command)
 Visualising the three principal components using a 3D scatter plot.
Visualising the three principal components using a 3D scatter plot.
Stick figures
# the XKCD package has a more flexible function for creating stick figures but perhaps for another day
# this draw.stick function is from
# https://github.com/EconometricsBySimulation/R-Graphics/blob/master/Stick-Figures/draw.stick.R
# Stick Man/Woman Generating Function
draw.stick <- function(x,y,scale=1,arms="down",
gender="male",lwd=3, clcol="white",
face="happy", linecol=gray(.3),
hat=NA) {
# clcol: color of clothes - any color
# scale: fize of figure
# x,y left bottom alignment of figure
# linecol: color of lines - any color
# lwd: line weight
# Arms: "down", "neutral", "up", "hip", "wave"
# Gender: "male", "female"
# Face: "happy", "sad", "annoyed", "surprised"
# Hat: plot hat T,F
# Set the figure scale, default it 1
s <- scale/100
# If is undefined then give the man a hat
if (is.na(hat)) hat<-(gender=="male")
require("plotrix")
# Draw Head
draw.ellipse(x+50*s,y+75*s,10*s,15*s,lwd=lwd, border=linecol)
if (face=="happy") {
# Draw eyes
draw.ellipse(x+46*s,y+77*s,2.5*s,2*s,lwd=lwd, border=linecol)
draw.ellipse(x+54*s,y+77*s,2.5*s,2*s,lwd=lwd, border=linecol)
# Draw mouth
draw.ellipse(x+50*s,y+72*s,6*s,8*s, segment = c(-160,-20),
lwd=lwd, border=linecol)
}
if (face=="sad") {
# Draw eyes
draw.ellipse(x+46*s,y+75*s,2*s,2*s,lwd=lwd, border=linecol)
draw.ellipse(x+54*s,y+75*s,2*s,2*s,lwd=lwd, border=linecol)
# Draw mouth
draw.ellipse(x+50*s,y+60*s,6*s,8*s, segment = c(140,40),
lwd=lwd, border=linecol)
}
if (face=="surprised") {
# Draw eyes
draw.ellipse(x+46*s,y+78*s,3*s,2*s,lwd=lwd, border=linecol)
draw.ellipse(x+54*s,y+78*s,3*s,2*s,lwd=lwd, border=linecol)
# Draw irises
# draw.ellipse(x+46*s,y+78*s,1*s,1*s,lwd=lwd, border=linecol)
# draw.ellipse(x+54*s,y+78*s,1*s,1*s,lwd=lwd, border=linecol)
# Draw mouth
draw.ellipse(x+50*s,y+65*s,3*s,4*s,
lwd=lwd, border=linecol)
}
if (face=="annoyed") {
# Draw mouth
lines(c(x+46*s,x+55*s), c(y+66*s,y+68*s),lwd=lwd, col=linecol)
# Draw eyes
draw.ellipse(x+46*s,y+76*s,2*s,2*s,lwd=lwd, border=linecol)
draw.ellipse(x+54*s,y+76*s,2*s,1*s,lwd=lwd, border=linecol)
}
# Draw torso
lines(c(x+50*s,x+50*s), c(y+35*s,y+60*s),lwd=lwd, col=linecol)
# Draw arms
if (arms=="down") {
lines(c(x+50*s,x+36*s), c(y+55*s,y+30*s),lwd=lwd, col=linecol) # Left
lines(c(x+50*s,x+64*s), c(y+55*s,y+30*s),lwd=lwd, col=linecol) # Right
}
if (arms=="neutral") {
lines(c(x+50*s,x+30*s), c(y+50*s,y+55*s),lwd=lwd, col=linecol) # Left
lines(c(x+50*s,x+70*s), c(y+50*s,y+55*s),lwd=lwd, col=linecol) # Right
}
if (arms=="up") {
lines(c(x+50*s,x+32*s), c(y+50*s,y+65*s),lwd=lwd, col=linecol) # Left
lines(c(x+50*s,x+68*s), c(y+50*s,y+65*s),lwd=lwd, col=linecol) # Right
}
if (arms=="hip") {
lines(c(x+50*s,x+37*s,x+48*s), c(y+56*s,y+47*s,y+40*s),lwd=lwd, col=linecol) # Left
lines(c(x+50*s,x+63*s,x+51*s), c(y+56*s,y+49*s,y+62*s),lwd=lwd, col=linecol) # Right
}
if (arms=="wave") {
lines(c(x+50*s,x+38*s,x+33*s), c(y+56*s,y+60*s,y+78*s),lwd=lwd, col=linecol) # Left
lines(c(x+50*s,x+63*s,x+52*s), c(y+56*s,y+47*s,y+40*s),lwd=lwd, col=linecol) # Right
}
# Draw male legs
if (gender=="male") {
lines(c(x+50*s,x+40*s), c(y+35*s,y+5*s),lwd=lwd, col=linecol)
lines(c(x+50*s,x+60*s), c(y+35*s,y+5*s),lwd=lwd, col=linecol)
}
# Draw female legs and dress
if (gender!="male") {
# Draw legs
lines(c(x+45*s,x+45*s), c(y+17*s,y+5*s),lwd=lwd, col=linecol)
lines(c(x+55*s,x+55*s), c(y+17*s,y+5*s),lwd=lwd, col=linecol)
# Draw dress
polygon(c(x+s*50,x+s*35,x+s*65),
c(y+s*40,y+s*17,y+s*17),
col=clcol, border=linecol,lwd=lwd)
}
# Draw hat
if (hat==T) polygon(
c(x+35*s,x+65*s,x+65*s,x+59*s,x+58*s,x+42*s,x+41*s,x+35*s),
c(y+84*s,y+84*s,y+86*s,y+86*s,y+91*s,y+91*s,y+86*s,y+86*s),
col=clcol, border=linecol,lwd=lwd)
}
frames <- 50
rename <- function(x){
if (x < 10) {
return(name <- paste('000',i,'plot.png',sep=''))
}
if (x < 100 && i >= 10) {
return(name <- paste('00',i,'plot.png', sep=''))
}
if (x >= 100) {
return(name <- paste('0', i,'plot.png', sep=''))
}
}
#loop through plots
for(i in 1:frames){
name <- rename(i)
#saves the plot as a .png file in the working directory
png(name)
# Arms: "down", "neutral", "up", "hip", "wave"
# Gender: "male", "female"
# Face: "happy", "sad", "annoyed", "surprised"
# Hat: plot hat T,F
a <- c('down','neutral','up','hip','wave')
f <- c('happy','sad','annoyed','surprised')
h <- c(TRUE,FALSE)
fs <- sample(f,1)
as <- sample(a,1)
hs <- sample(h,1)
plot(c(.25,.75),
c(0,1),
type="n",
main=paste(fs, as, hs)
)
draw.stick(0,0, face=fs, gender="male", arms=as, hat=hs)
dev.off()
}
#run ImageMagick
my_command <- 'convert *.png -delay 5 -loop 0 stick_figure.gif'
system(my_command)

This work is licensed under a Creative Commons
Attribution 4.0 International License.

Wow, great post. I too watched in horror as classmates defiled text books. I recently wanted to highlight assigned homework numbers in a text book. I considered how to do this for at least 10 min before making the slightest of dots next to the numbers. 😉
Back to topic, I love the independent filtering gif. I might add to that a histogram showing number of deg at each filter value with the bar color changing/following as the value changes and a Venn showing overlap of degs between current and baseline (no filtering).
Fastest comment I’ve ever gotten 🙂 And thanks for the great suggestion!
It took me a while to find a way to overlay text on the 3D PCA plot. The package vignette, http://cran.r-project.org/web/packages/scatterplot3d/vignettes/s3d.pdf, shows how to do it on page 18. To make it work in Dave’s example, simply use this code:
#loop through plots
for(i in 1:frames){
name <- rename(i)
#saves the plot as a .png file in the working directory
png(name)
s3d <- scatterplot3d(p$x[,1:3],
main=paste("Angle", i),
angle=i,
pch=19,
cex.symbols=0.5,
color=my_col)
# Overlaying text
s3d.coords <- s3d$xyz.convert(p$x[,1:3])
text(s3d.coords$x, s3d.coords$y, labels=colnames(iris[, 1:4]), pos=2, offset=0.5, cex=0.7)
dev.off()
}
Obviously, it works best when the plot is not as crowded.
And, at the end of the example for the 3D PCA plot, I'd add a cleanup command
system("rm *.png")
With R 3.4.2 and ImageMagick 7.0.7-28 Q16 x64, you’ll need the following call, instead of system():
system2(“magick”, c(“convert”, “*.png”, “-delay 3”, “-loop 0”, “animation.gif”))
Thanks, Dave for a very cool set of animation examples.
I’m just getting started into that world & tried adapting you’re example to iterate through
a dataframe 1 row at a time to produce a basic line plot *unsuccessfully*, though.
* All 50 png files created include the plot of the entire data series ?
Do I need to add a sleep() cmd to the iterative plot generation?
# ——
frames
I haven’t had a chance to check out gganimate (https://github.com/thomasp85/gganimate) but perhaps it might be easier to follow than my blog post.