The UCSC Genome Browser provides multiple alignments of 46 vertebrate species and conveniently provides them for download. The multiple alignments show regions of sequence conservation among vertebrates. For more information see http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=cons46way. The multiple alignments are stored as Multiple Alignment Files and there are Perl and Python packages that parse them. The MAF format is quite straight forward and here I convert the MAF files into a BED file for use with intersectBed from the BEDTools suite.
multiz46way
#!/bin/bash wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr1.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr2.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr3.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr4.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr5.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr6.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr7.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr8.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr9.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr10.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr11.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr12.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr13.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr14.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr15.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr16.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr17.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr18.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr19.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr20.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr21.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chr22.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chrM.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chrX.maf.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/multiz46way/maf/chrY.maf.gz
Convert the MAF files into BED:
#!/usr/bin/perl
use strict;
use warnings;
=head
##maf version=1 scoring=autoMZ.v1
a score=1189.000000
s hg19.chr22 16050000 8 + 51304566 GATCTGAT
s panTro2.chrUn 7680399 8 + 58616431 gatctgat
q panTro2.chrUn 99999999
i panTro2.chrUn N 0 C 0
s choHof1.scaffold_39500 681 6 + 15110 --TCAGGG
q choHof1.scaffold_39500 --999999
i choHof1.scaffold_39500 N 0 N 0
s dasNov2.scaffold_31225 17898 6 - 18719 --TCAGGG
q dasNov2.scaffold_31225 --999999
i dasNov2.scaffold_31225 N 0 N 0
=cut
my $usage = "Usage: $0 <infile.maf>\n";
my $infile = shift or die $usage;
if ($infile =~ /\.gz$/){
open(IN,'-|',"zcat $infile") || die "Could not open $infile: $!\n";
} else {
open(IN,'<',$infile) || die "Could not open $infile: $!\n";
}
my @alignment = ();
my $score = '';
while(<IN>){
chomp;
next if /^#/;
if (/^a/){
if (/score=(-*\d+)\./){
$score = $1;
} else {
die "Could not parse score on line $.: $_\n";
}
}
elsif (/^s/){
push(@alignment,$_);
} elsif (/^$/){
#first should always be hg19
my $homologous_region = scalar(@alignment);
my ($junk,$src,$start,$size,$strand,$srcSize,$text) = split(/\s+/,$alignment[0]);
my ($genome,$chr) = split(/\./,$src);
if ($strand eq '+'){
my $id = join("_",$chr,$start,$start+$size,$homologous_region);
print join("\t",$chr,$start,$start+$size,$id,$score,$strand),"\n";
} elsif ($strand eq '-'){
my $id = join("_",$chr,$start-$size,$start,$homologous_region);
print join("\t",$chr,$start-$size,$start,$id,$score,$strand),"\n";
} else {
die "Unrecognised strand: $strand on line $alignment[0]\n";
}
#for (my $i=0; $i<scalar(@alignment); ++$i){
#}
@alignment = ();
$score = '';
}
}
close(IN);
exit(0);
__END__
BASH scripting to generate all BED files:
for file in `ls *.gz`; do base=`basename $file .maf.gz`; echo $base; maf.pl $file > ${base}.bed; done
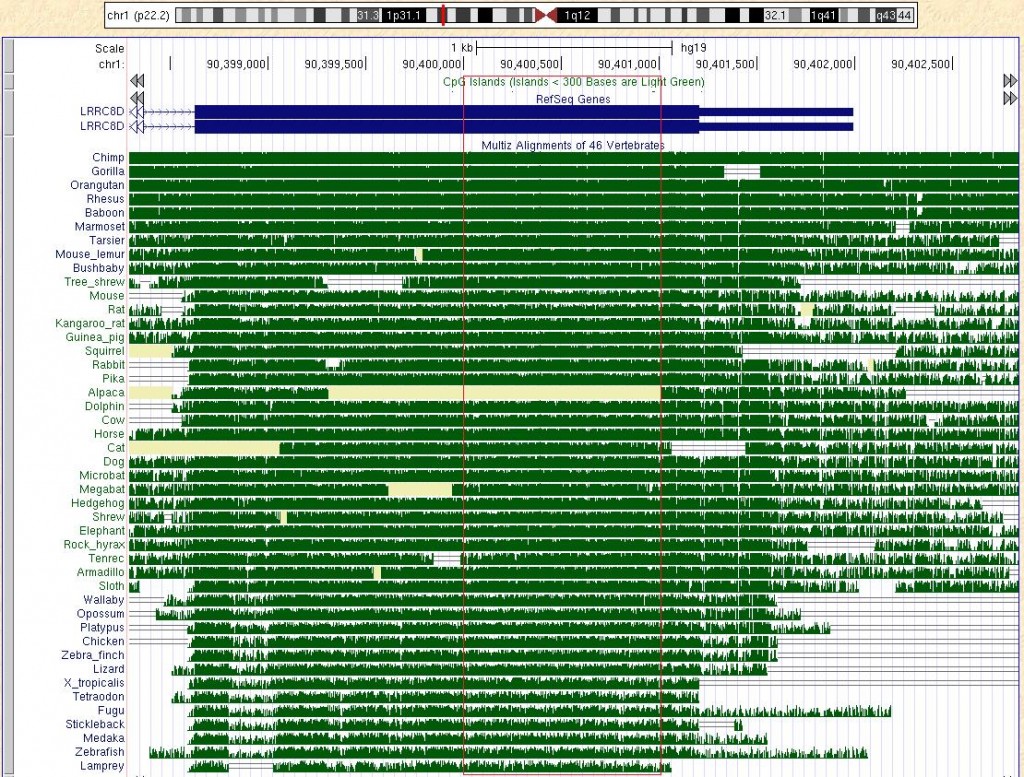
By sorting on the score column of chr1.bed, we can find the most conserved region on chromosome 1 of hg19 (in the red rectangle), which happens to be the 3' tail of the LRRC8D gene:
chr1 90400115 90401012 chr1_90400115_90401012_45 61991922 +
phyloP46way and phastCons46way
There are also two methods that show measurements of sequence conservation among the 46 vertebrate genomes. Quoting the UCSC Genome Browser:
PhastCons (which has been used in previous Conservation tracks) is a hidden Markov model-based method that estimates the probability that each nucleotide belongs to a conserved element, based on the multiple alignment. It considers not just each individual alignment column, but also its flanking columns. By contrast, phyloP separately measures conservation at individual columns, ignoring the effects of their neighbours. As a consequence, the phyloP plots have a less smooth appearance than the phastCons plots, with more "texture" at individual sites. The two methods have different strengths and weaknesses. PhastCons is sensitive to "runs" of conserved sites, and is therefore effective for picking out conserved elements. PhyloP, on the other hand, is more appropriate for evaluating signatures of selection at particular nucleotides or classes of nucleotides (e.g., third codon positions, or first positions of miRNA target sites).
If I turn on the PhastCons and phyloP scores on the UCSC Genome Browser for the region I highlighted above:
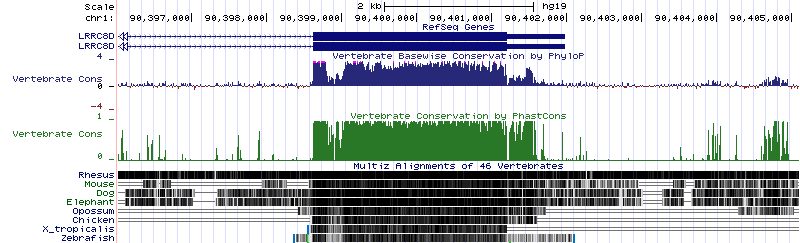 Concordant agreement (concordance according to my eye) between the two methods showing a long stretch of sequence conservation in the tail of the LRRC8D gene.
Concordant agreement (concordance according to my eye) between the two methods showing a long stretch of sequence conservation in the tail of the LRRC8D gene.
These scores are available as fixedStep wiggle plots.
Download the phyloP46way files:
wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr1.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr2.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr3.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr4.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr5.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr6.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr7.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr8.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr9.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr10.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr11.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr12.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr13.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr14.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr15.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr16.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr17.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr18.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr19.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr20.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr21.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chr22.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chrM.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chrX.phyloP46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phyloP46way/vertebrate/chrY.phyloP46way.wigFix.gz
Download the phastCons46way files:
wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr1.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr2.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr3.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr4.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr5.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr6.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr7.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr8.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr9.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr10.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr11.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr12.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr13.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr14.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr15.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr16.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr17.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr18.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr19.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr20.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr21.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chr22.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chrM.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chrX.phastCons46way.wigFix.gz wget http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way/vertebrate/chrY.phastCons46way.wigFix.gz
To create a more readable format of these files (but using up more space), download the wigToBigWig and the bigWigToBedGraph files from the UCSC Genome Browser's list of executables and convert them to the bedGraph format.
#convert to bigWig
for file in `ls *.gz`;
do base=`basename $file .wigFix.gz`;
echo $file;
wigToBigWig $file ~/davetang/chrom_size/hg19_chrom_info.txt ${base}.bw;
done
#convert to bedGraph
for file in `ls *.bw`;
do base=`basename $file .bw`;
echo $file;
bigWigToBedGraph $file $base.bedGraph;
done
rm *.bw *.wigFix.gz
Finding conserved intergenic regions
To find sequence conservation within intergenic regions, firstly calculate the intergenic regions. Let's look at chr22:
#the gencode_v17_intergenic.bed file was created following the link above
cat gencode_v17_intergenic.bed | grep chr22 > gencode_v17_intergenic_chr22.bed
cat gencode_v17_intergenic_chr22.bed | wc
746 2238 17897
#create 75 bp non-overlapping windows
bedtools makewindows -b gencode_v17_intergenic_chr22.bed -w 75 > gencode_v17_intergenic_chr22_75_window.bed
cat gencode_v17_intergenic_chr22_75_window.bed | wc
381936 1145808 8870165
#intersect the windows to the bedGraph file we created above
bedtools intersect -a gencode_v17_intergenic_chr22_75_window.bed -b ~/ucsc/phastCons46way/vertebrate/chr22.phastCons46way.bedGraph -wo > gencode_v17_intergenic_chr22_75_window_phastcons46way.tsv
head gencode_v17_intergenic_chr22_75_window_phastcons46way.tsv
chr22 16050000 16050075 chr22 16050000 16050001 0.093 1
chr22 16050000 16050075 chr22 16050001 16050002 0.086 1
chr22 16050000 16050075 chr22 16050002 16050003 0.079 1
chr22 16050000 16050075 chr22 16050003 16050004 0.068 1
chr22 16050000 16050075 chr22 16050004 16050005 0.05 1
chr22 16050000 16050075 chr22 16050005 16050006 0.045 1
chr22 16050000 16050075 chr22 16050006 16050007 0.035 1
chr22 16050000 16050075 chr22 16050007 16050008 0.032 1
chr22 16050000 16050075 chr22 16050008 16050009 0.034 1
chr22 16050000 16050075 chr22 16050009 16050011 0.035 2
#sum the phastCons scores per window
cat gencode_v17_intergenic_chr22_75_window_phastcons46way.tsv | groupBy -g 1,2,3 -c 7 -o sum | head
chr22 16050000 16050075 4.61099999999999976552
chr22 16050075 16050150 10.1810000000000009379
chr22 16050150 16050225 6.27699999999999658229
chr22 16050225 16050300 5.73000000000000042633
chr22 16050300 16050375 0.675000000000000155431
chr22 16050375 16050450 0.744000000000000327738
chr22 16050450 16050525 0.287000000000000088374
chr22 16050525 16050600 0.620000000000000106581
chr22 16050600 16050675 0.186000000000000054179
chr22 16050675 16050750 0.543000000000000038192
#find the most conserved block
cat gencode_v17_intergenic_chr22_75_window_phastcons46way.tsv | groupBy -g 1,2,3 -c 7 -o sum | sort -k4rn | head
chr22 22557650 22557725 68.5529999999999830607
chr22 48699552 48699627 67.7090000000000031832
chr22 20361407 20361482 67.5860000000000127329
chr22 48648171 48648246 67.550999999999973511
chr22 18705182 18705257 67.5360000000000013642
chr22 49308598 49308673 67.470000000000013074
chr22 22979374 22979449 67.4459999999999837428
chr22 48648246 48648321 67.354000000000013415
chr22 21012303 21012378 67.2660000000000053433
chr22 49308673 49308748 67.1859999999999928377
 Visualisation of the most conserved window (chr22:22557650-22557725) on the UCSC Genome Browser.
Visualisation of the most conserved window (chr22:22557650-22557725) on the UCSC Genome Browser.

This work is licensed under a Creative Commons
Attribution 4.0 International License.