I've seen talks over the years where the speaker shows a bar chart with the number of articles in PubMed that contain a certain keyword and tallied per year. In most of the cases the speaker was trying to illustrate the growing number of articles that contain the keyword. Here I try to do the same by querying PubMed using R.
#install the RISmed package
install.packages("RISmed")
library(RISmed)
#now let's look up this dude called Dave Tang
res <- EUtilsSummary('dave tang', type='esearch', db='pubmed')
summary(res)
Query:
Tang, Dave[Full Author Name]
Result count: 10
#what are the PubMed ids for the Author Dave Tang?
QueryId(res)
[1] "23180801" "22976001" "22722852" "21888672" "21386911" "20510229" "19648138" "19501082" "19393063"
[10] "19270757"
#limit by date
res2 <- EUtilsSummary('dave tang', type='esearch', db='pubmed', mindate='2012', maxdate='2012')
summary(res2)
Query:
Tang, Dave[Full Author Name] AND 2012[EDAT] : 2012[EDAT]
Result count: 3
#three publications in 2012
QueryId(res2)
[1] "23180801" "22976001" "22722852"
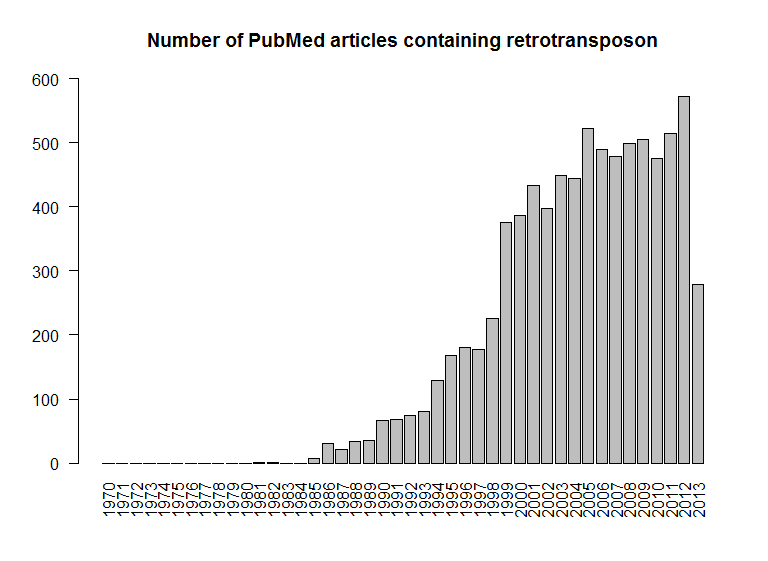
I'm interested in the number of publications containing the word "retrotransposon" that are in PubMed, tallied per year, from 1970 (the year the discovery of reverse transcriptase was published) to 2013.
#first how many total articles containing retrotransposon
res3 <- EUtilsSummary('retrotransposon', type='esearch', db='pubmed')
summary(res3)
Query:
"retroelements"[MeSH Terms] OR "retroelements"[All Fields] OR "retrotransposon"[All Fields]
Result count: 8123
#if you only want the number of articles
QueryCount(res3)
[1] 8123
#tally each year beginning at 1970
#In order not to overload the E-utility servers, NCBI recommends that users post no more than three
#URL requests per second and limit large jobs to either weekends or between 9:00 PM and 5:00 AM
#Eastern time during weekdays. Failure to comply with this policy may result in an IP address being
#blocked from accessing NCBI.
tally <- array()
x <- 1
for (i in 1970:2013){
Sys.sleep(1)
r <- EUtilsSummary('retrotransposon', type='esearch', db='pubmed', mindate=i, maxdate=i)
tally[x] <- QueryCount(r)
x <- x + 1
}
names(tally) <- 1970:2013
max(tally)
[1] 573
barplot(tally, las=2, ylim=c(0,600), main="Number of PubMed articles containing retrotransposon")
How about the word "transposon"?
transposon <- array()
x <- 1
for (i in 1970:2013){
Sys.sleep(1)
r <- EUtilsSummary('transposon', type='esearch', db='pubmed', mindate=i, maxdate=i)
transposon[x] <- QueryCount(r)
x <- x + 1
}
names(transposon) <- 1970:2013
max(transposon)
[1] 1634
barplot(transposon, las=2, ylim=c(0,2000), main="Number of PubMed articles containing transposon")
Is there an upward trend for any keyword, due to the increase in the number of journals and database entries?
trna <- array()
x <- 1
for (i in 1970:2013){
Sys.sleep(1)
r <- EUtilsSummary('trna', type='esearch', db='pubmed', mindate=i, maxdate=i)
trna[x] <- QueryCount(r)
x <- x + 1
}
names(trna) <- 1970:2013
max(trna)
[1] 2015
barplot(trna, las=2, ylim=c(0,2000), main="Number of PubMed articles containing trna")
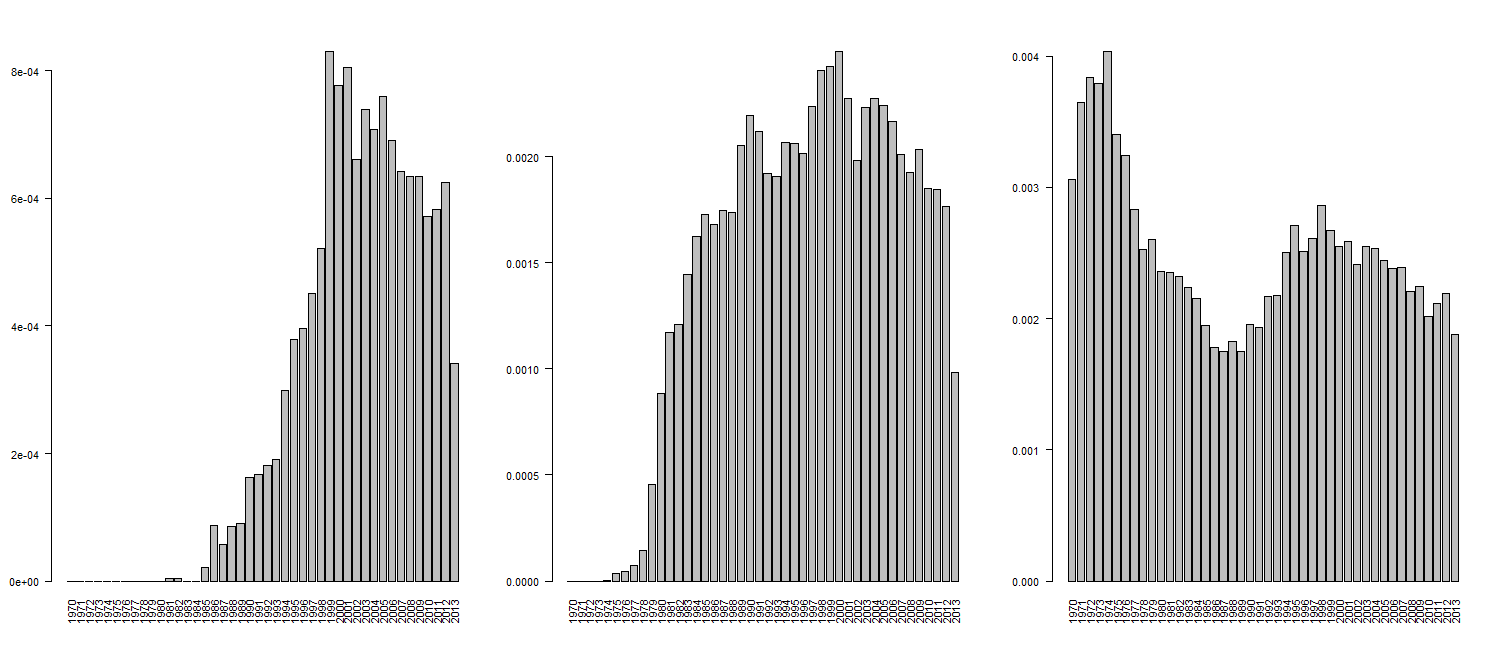
Normalising by total article counts
To get the total number of articles for a given year, just use an empty query.
test <- EUtilsSummary('', type='esearch', db='pubmed', mindate=1970, maxdate=1970)
summary(test)
Query:
1970[EDAT] : 1970[EDAT]
Result count: 218690
#number of articles each year
total <- array()
x <- 1
for (i in 1970:2013){
Sys.sleep(1)
r <- EUtilsSummary('', type='esearch', db='pubmed', mindate=i, maxdate=i)
total[x] <- QueryCount(r)
x <- x + 1
}
names(total) <- 1970:2013
max(total)
[1] 917657
barplot(total, las=2, ylim=c(0,1000000), main="Number of PubMed articles each year")
 The dip in 1997 is a consequence of the 1997 Asian financial crisis?
The dip in 1997 is a consequence of the 1997 Asian financial crisis?
Now to normalise the previous searches:
tally_norm <- tally / total transposon_norm <- transposon / total trna_norm <- trna / total par(mfrow=c(1,3)) barplot(tally_norm, las=2) barplot(transposon_norm, las=2) barplot(trna_norm, las=2) #reset par(mfrow=c(1,1))
Conclusions
So there you have it, querying PubMed using R via the CRAN package RISmed. More complicated queries can be built and for those interested have a look at the manual.

This work is licensed under a Creative Commons
Attribution 4.0 International License.



Very very interesting!!!
🙂
does pubmed keep track of citations per article per year?
I’m not sure about that. All I know is that Google Scholar can provide you with the number of citations per year for articles that have your name on it.
awsome!
Very nice! However, why instead of running several queries don’t you just use EUtilsGet on the EUtilsSummary result? This way you just run one query and can then filter the result offline.
Thanks for the comment. That sounds much more desirable; I’ll update the post accordingly later. Thanks!
Thank you for this very clear and interesting example of RisMed’s usage
Do you use windows for running this code? I tried using on mac and I got the error regarding “setInternet2(TRUE)”. It has been resolved for windows only but not for mac.
Works for me.
library(RISmed) res <- EUtilsSummary('dave tang', type='esearch', db='pubmed') summary(res) Query: Tang, Dave[Full Author Name] Result count: 16 sessionInfo() R version 3.4.3 (2017-11-30) Platform: x86_64-apple-darwin15.6.0 (64-bit) Running under: macOS High Sierra 10.13.2 Matrix products: default BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib locale: [1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8 attached base packages: [1] stats graphics grDevices utils datasets methods base other attached packages: [1] RISmed_2.1.7 BiocInstaller_1.28.0 loaded via a namespace (and not attached): [1] compiler_3.4.3 tools_3.4.3 yaml_2.1.15Haven’t used the package myself, just check your post. I observed that the first argument in the EUtilesSummary function is a single entry term; however in practice we use terms connected by Booleans to query the database. So, I’m curious if the function can take a query containing multiple Booleans or proximity operators?